Cet article propose de multiples solutions pour créer un magnifique diagramme de Venn avec R ou RStudio. Les packages R suivants seront illustrés : ggvenn, ggVenDiagram, VennDiagram et gplots.
Sommaire:
Créer une donnée de démonstration
set.seed(20190708)
genes <- paste("gene",1:1000,sep="")
x <- list(
A = sample(genes,300),
B = sample(genes,525),
C = sample(genes,440),
D = sample(genes,350)
)Utilisation du package R ggvenn
Ce package est une extension de ggplot2. Une couleur peut être attribuée à chaque groupe.
Installation:
if (!require(devtools)) install.packages("devtools")
devtools::install_github("yanlinlin82/ggvenn")Utilisation:
library(ggvenn)
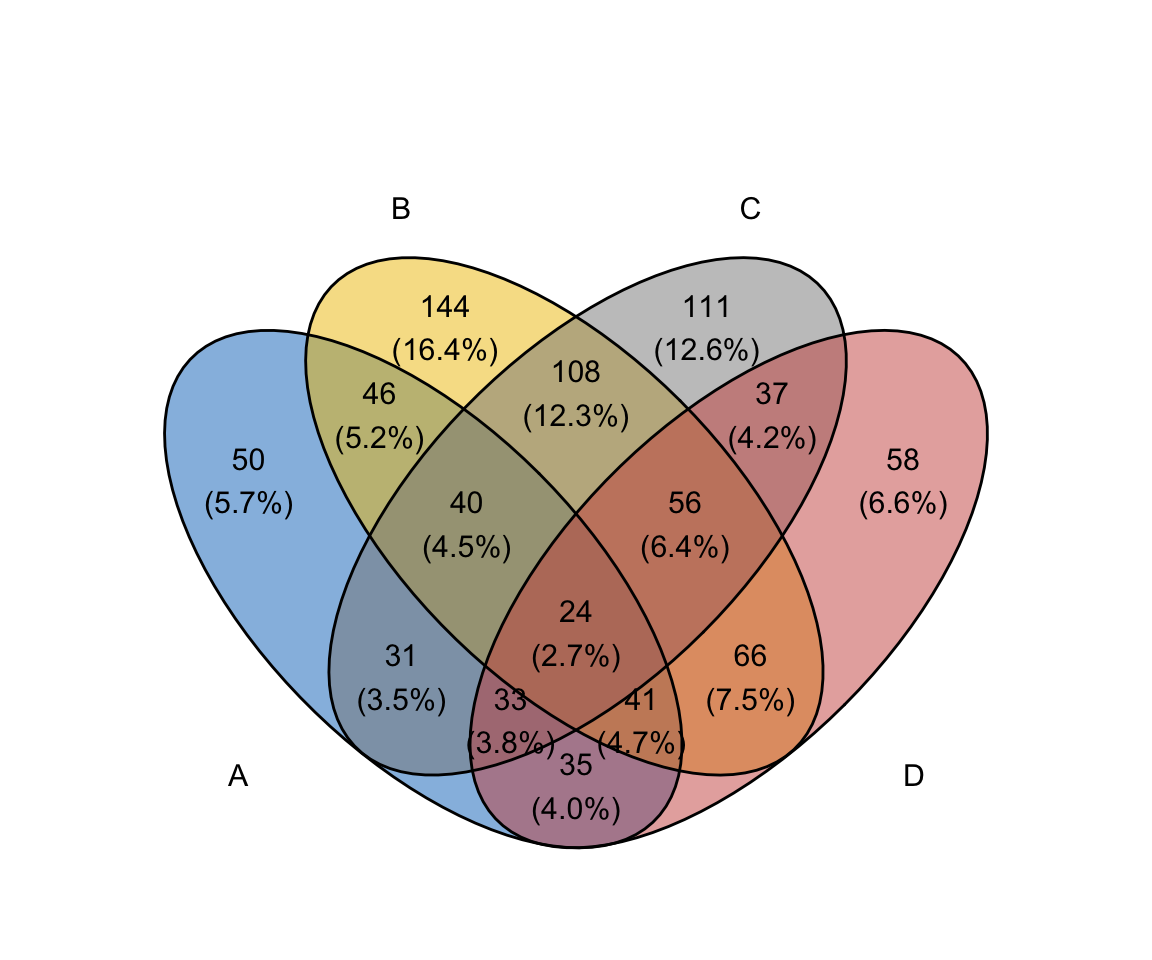
ggvenn(
x,
fill_color = c("#0073C2FF", "#EFC000FF", "#868686FF", "#CD534CFF"),
stroke_size = 0.5, set_name_size = 4
)
Plus d’informations ici : Beau diagramme de Venn GGPlot avec R
Utilisation du package R ggVennDiagram
Ce package est une extension de ggplot2. Le package ggVennDiagram fait correspondre la couleur de remplissage de chaque région à une quantité, ce qui nous permet d’observer visuellement les différences entre les différentes parties.
Installation:
if (!require(devtools)) install.packages("devtools")
devtools::install_github("gaospecial/ggVennDiagram")Utilisation:
library("ggVennDiagram")
ggVennDiagram(x, label_alpha = 0)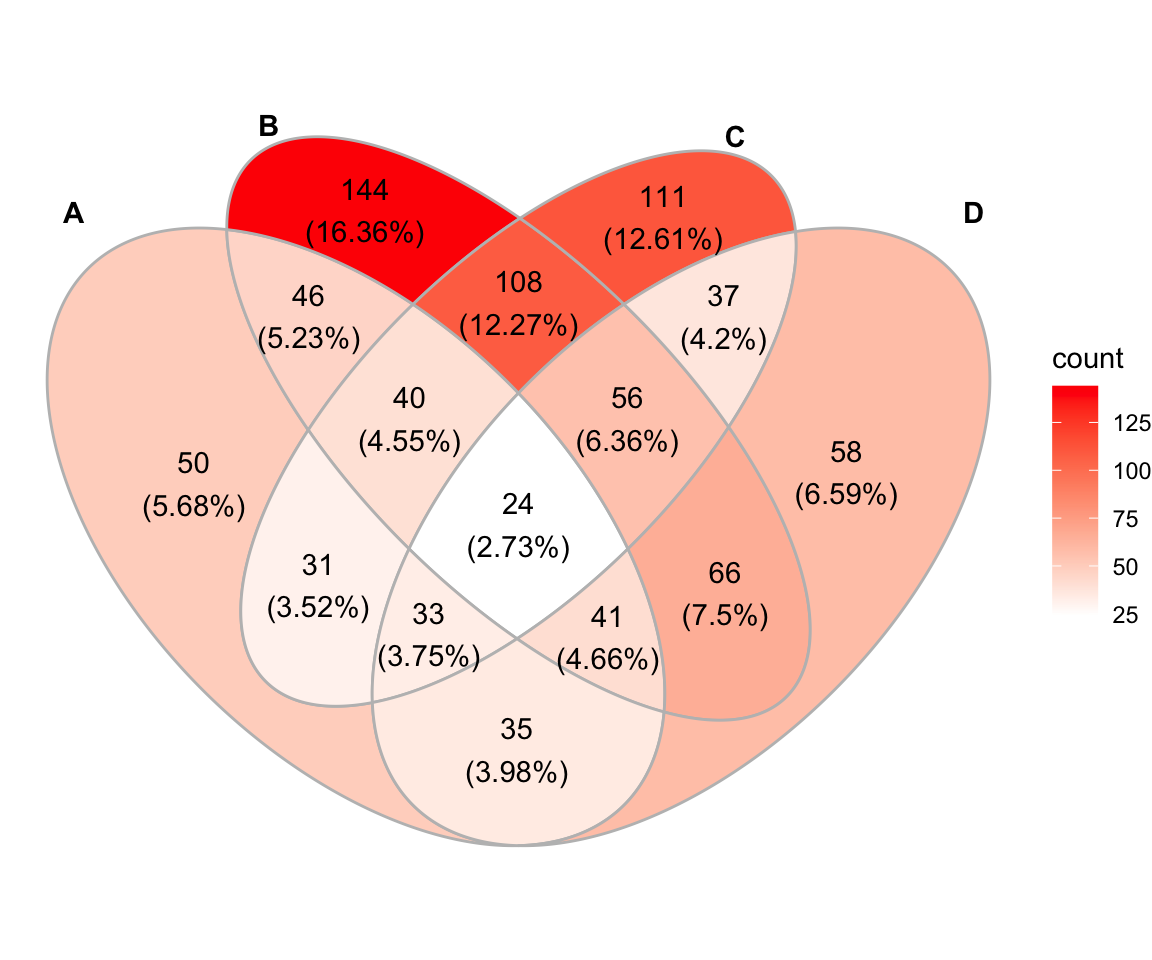
Plus d’informations ici : Beau diagramme de Venn GGPlot avec R
Utilisation du package R VennDiagram
Installation:
install.packages("VennDiagram")`Utilisation:
Créer un diagramme de Venn et l’enregistrer dans un fichier. La fonction venn.diagram() prend une liste et crée un fichier contenant un diagramme de Venn de qualité publication.
library(VennDiagram)
venn.diagram(x, filename = "venn-4-dimensions.png")Afficher le graphique directement dans R:
# Fonction d'aide pour afficher le diagramme de Venn
display_venn <- function(x, ...){
library(VennDiagram)
grid.newpage()
venn_object <- venn.diagram(x, filename = NULL, ...)
grid.draw(venn_object)
}# Diagramme de Venn en quatre dimensions
display_venn(x)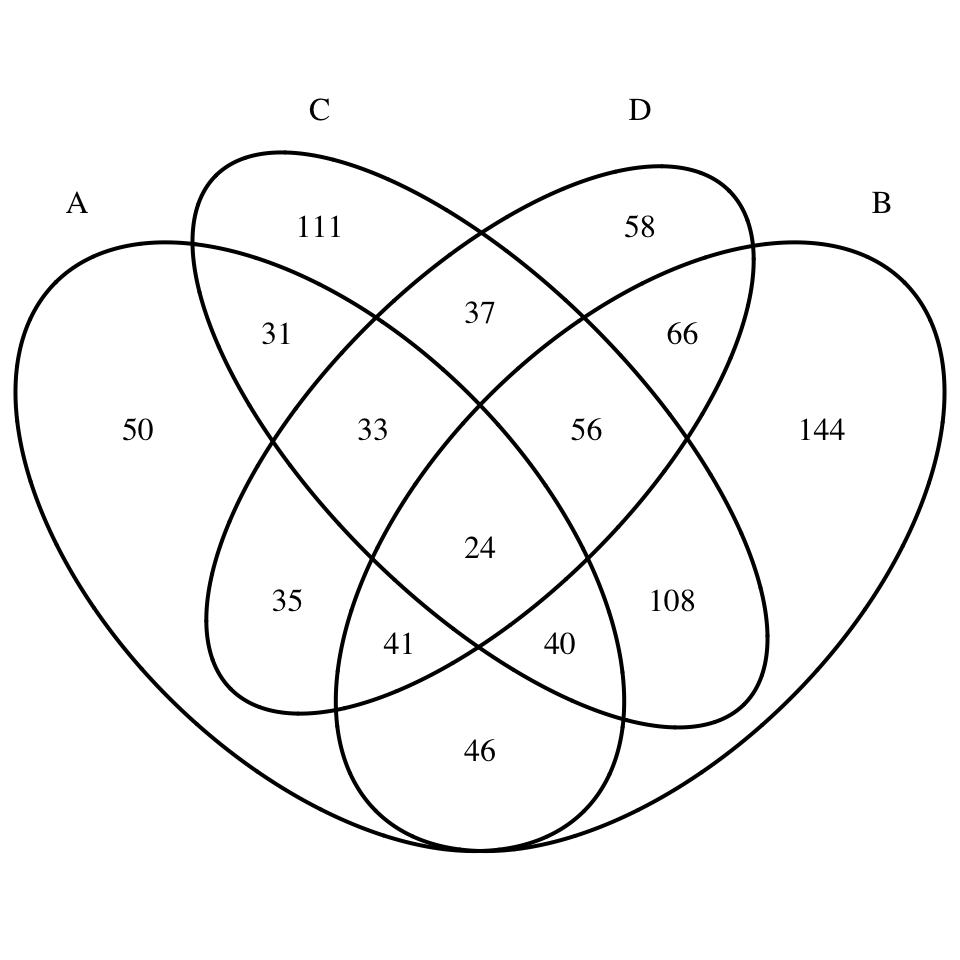
# Diagramme de Venn en trois dimensions
display_venn(x[1:3])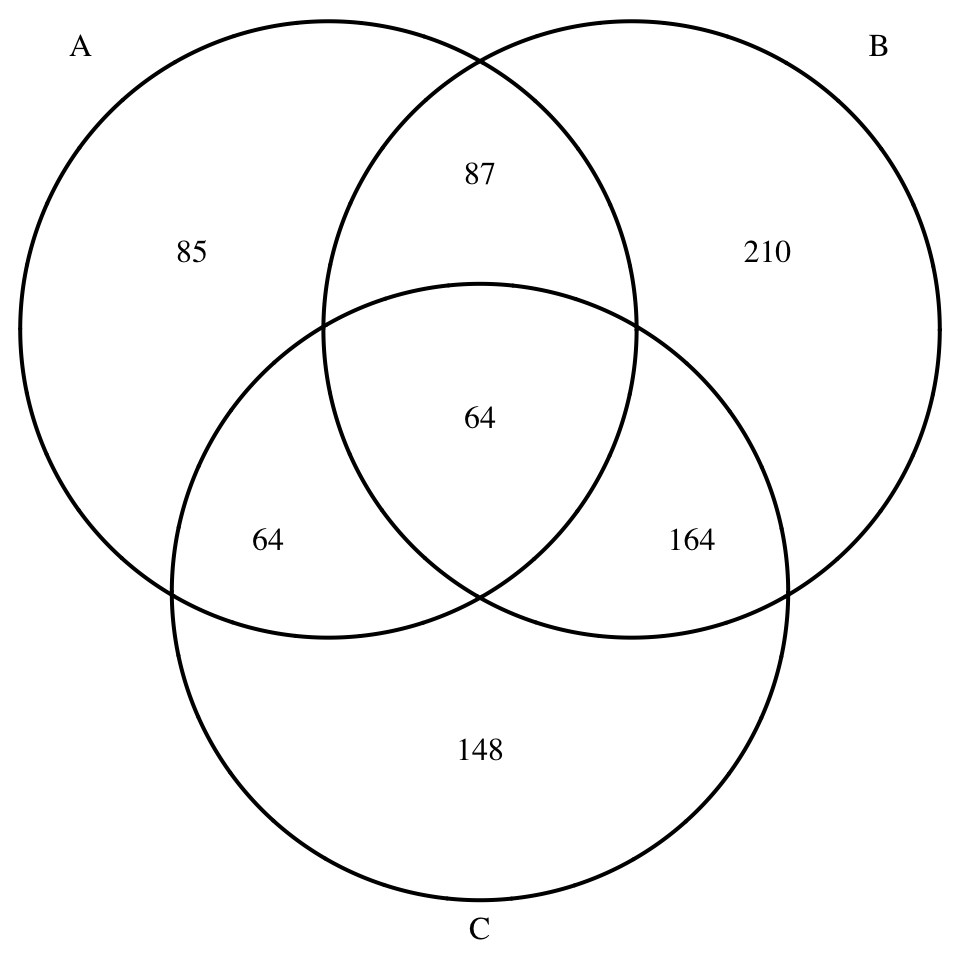
# Changer les noms de catégories
# Changer la couleur de remplissage
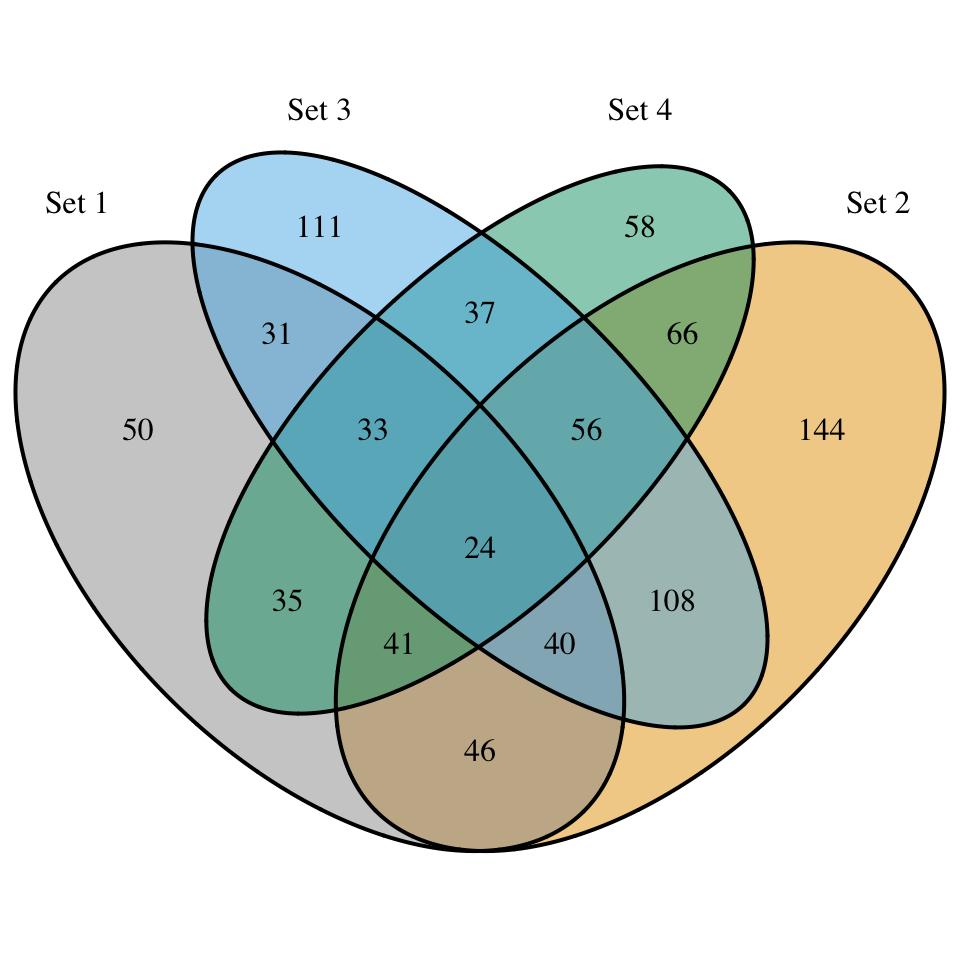
display_venn(
x,
category.names = c("Set 1" , "Set 2 " , "Set 3", "Set 4"),
fill = c("#999999", "#E69F00", "#56B4E9", "#009E73")
)
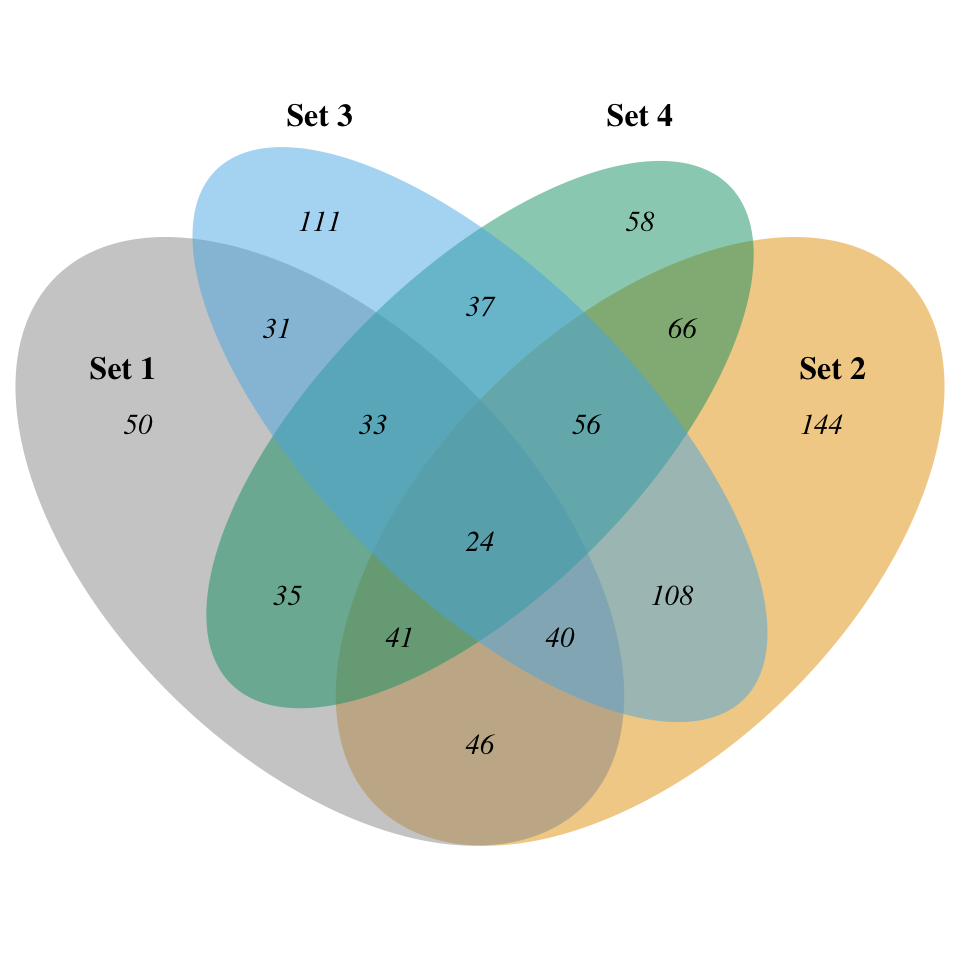
# Personnalisation supplémentaire
display_venn(
x,
category.names = c("Set 1" , "Set 2 " , "Set 3", "Set 4"),
# Cercles
lwd = 2,
lty = 'blank',
fill = c("#999999", "#E69F00", "#56B4E9", "#009E73"),
# Nombres
cex = .9,
fontface = "italic",
# Noms des groupes
cat.cex = 1,
cat.fontface = "bold",
cat.default.pos = "outer",
cat.dist = c(0.055, 0.055, 0.1, 0.1)
)
Utilisation du package R gplots
Installation:
install.package("gplots")`Utilisation:
library(gplots)
v.table <- venn(x)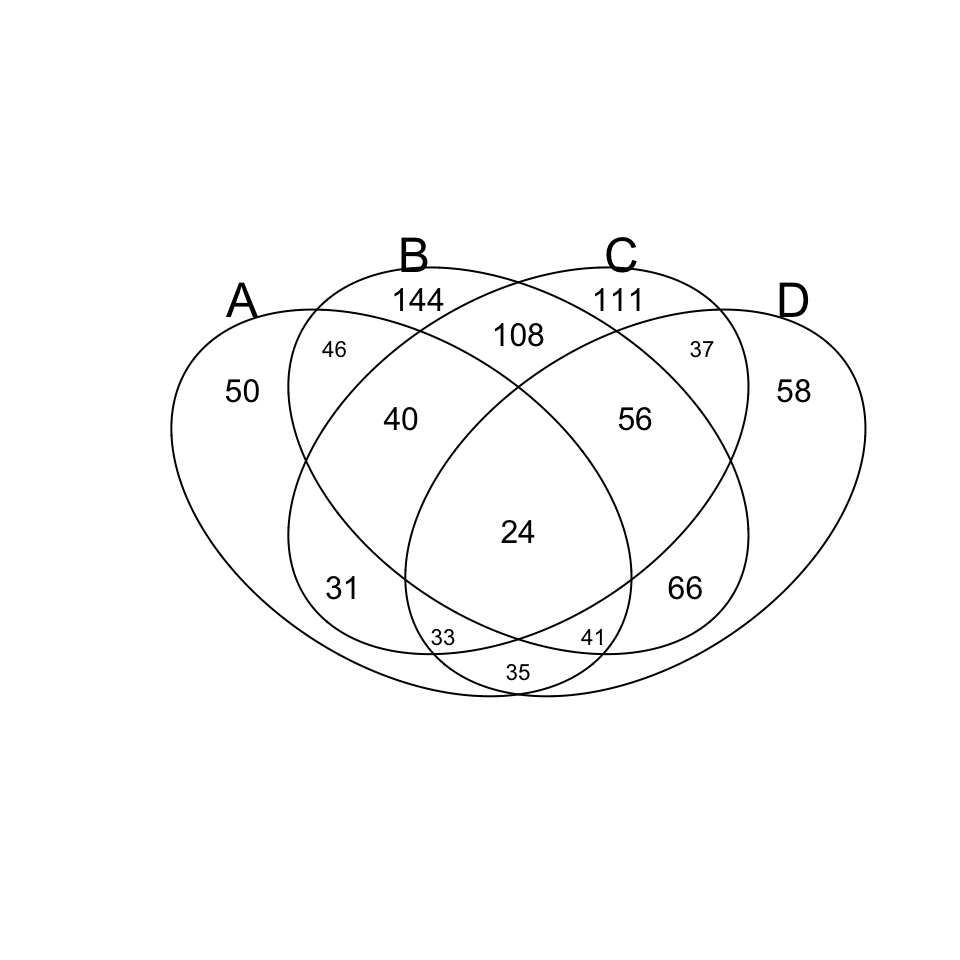
Accès aux données relatives aux intersections:
print(v.table)## num A B C D
## 0000 0 0 0 0 0
## 0001 58 0 0 0 1
## 0010 111 0 0 1 0
## 0011 37 0 0 1 1
## 0100 144 0 1 0 0
## 0101 66 0 1 0 1
## 0110 108 0 1 1 0
## 0111 56 0 1 1 1
## 1000 50 1 0 0 0
## 1001 35 1 0 0 1
## 1010 31 1 0 1 0
## 1011 33 1 0 1 1
## 1100 46 1 1 0 0
## 1101 41 1 1 0 1
## 1110 40 1 1 1 0
## 1111 24 1 1 1 1
## attr(,"intersections")
## attr(,"intersections")$A
## [1] "gene835" "gene910" "gene866" "gene785" "gene532" "gene385" "gene201" "gene74" "gene941" "gene401" "gene642"
## [12] "gene299" "gene819" "gene245" "gene834" "gene700" "gene422" "gene176" "gene643" "gene852" "gene584" "gene560"
## [23] "gene638" "gene164" "gene122" "gene419" "gene521" "gene363" "gene724" "gene160" "gene988" "gene328" "gene452"
## [34] "gene236" "gene707" "gene865" "gene342" "gene165" "gene572" "gene565" "gene206" "gene663" "gene107" "gene620"
## [45] "gene917" "gene298" "gene402" "gene840" "gene992" "gene225"
##
## attr(,"intersections")$B
## [1] "gene129" "gene150" "gene307" "gene73" "gene651" "gene40" "gene722" "gene374" "gene779" "gene772"
## [11] "gene574" "gene355" "gene970" "gene429" "gene979" "gene656" "gene208" "gene274" "gene252" "gene585"
## [21] "gene662" "gene535" "gene709" "gene589" "gene964" "gene569" "gene113" "gene851" "gene111" "gene308"
## [31] "gene18" "gene309" "gene977" "gene530" "gene929" "gene720" "gene83" "gene256" "gene217" "gene287"
## [41] "gene596" "gene776" "gene573" "gene99" "gene470" "gene689" "gene44" "gene473" "gene380" "gene246"
## [51] "gene774" "gene799" "gene621" "gene134" "gene659" "gene947" "gene322" "gene397" "gene664" "gene266"
## [61] "gene556" "gene210" "gene272" "gene389" "gene84" "gene771" "gene55" "gene349" "gene51" "gene137"
## [71] "gene462" "gene106" "gene353" "gene801" "gene943" "gene297" "gene678" "gene546" "gene617" "gene289"
## [81] "gene480" "gene361" "gene913" "gene875" "gene948" "gene777" "gene788" "gene743" "gene199" "gene879"
## [91] "gene229" "gene951" "gene447" "gene175" "gene715" "gene942" "gene668" "gene698" "gene559" "gene812"
## [101] "gene52" "gene337" "gene413" "gene757" "gene472" "gene526" "gene876" "gene567" "gene813" "gene147"
## [111] "gene685" "gene114" "gene614" "gene427" "gene508" "gene247" "gene540" "gene986" "gene241" "gene731"
## [121] "gene591" "gene279" "gene120" "gene346" "gene403" "gene333" "gene922" "gene135" "gene200" "gene603"
## [131] "gene914" "gene820" "gene492" "gene19" "gene978" "gene804" "gene655" "gene332" "gene958" "gene903"
## [141] "gene564" "gene989" "gene386" "gene151"
##
## attr(,"intersections")$C
## [1] "gene169" "gene390" "gene265" "gene581" "gene250" "gene634" "gene188" "gene319" "gene994" "gene366"
## [11] "gene213" "gene610" "gene661" "gene654" "gene453" "gene935" "gene193" "gene251" "gene243" "gene545"
## [21] "gene178" "gene411" "gene737" "gene953" "gene902" "gene557" "gene691" "gene221" "gene920" "gene495"
## [31] "gene474" "gene543" "gene695" "gene100" "gene686" "gene383" "gene125" "gene608" "gene278" "gene59"
## [41] "gene895" "gene828" "gene288" "gene611" "gene441" "gene396" "gene93" "gene448" "gene428" "gene367"
## [51] "gene438" "gene459" "gene579" "gene395" "gene248" "gene867" "gene45" "gene29" "gene437" "gene77"
## [61] "gene70" "gene484" "gene633" "gene85" "gene854" "gene490" "gene177" "gene426" "gene887" "gene550"
## [71] "gene923" "gene792" "gene412" "gene212" "gene590" "gene292" "gene795" "gene30" "gene159" "gene304"
## [81] "gene471" "gene62" "gene790" "gene28" "gene770" "gene533" "gene575" "gene173" "gene832" "gene860"
## [91] "gene683" "gene878" "gene818" "gene944" "gene789" "gene357" "gene391" "gene888" "gene919" "gene796"
## [101] "gene631" "gene82" "gene733" "gene732" "gene197" "gene368" "gene909" "gene816" "gene843" "gene765"
## [111] "gene898"
##
## attr(,"intersections")$D
## [1] "gene738" "gene321" "gene149" "gene938" "gene487" "gene145" "gene783" "gene15" "gene871" "gene475" "gene528"
## [12] "gene706" "gene130" "gene570" "gene43" "gene489" "gene640" "gene161" "gene293" "gene966" "gene171" "gene498"
## [23] "gene926" "gene701" "gene632" "gene649" "gene853" "gene702" "gene869" "gene831" "gene392" "gene694" "gene271"
## [34] "gene650" "gene880" "gene906" "gene541" "gene538" "gene420" "gene140" "gene677" "gene124" "gene644" "gene233"
## [45] "gene868" "gene152" "gene800" "gene12" "gene496" "gene39" "gene726" "gene674" "gene226" "gene379" "gene916"
## [56] "gene602" "gene857" "gene523"
##
## attr(,"intersections")$`A:B`
## [1] "gene168" "gene301" "gene86" "gene133" "gene786" "gene235" "gene209" "gene205" "gene451" "gene372" "gene794"
## [12] "gene889" "gene838" "gene604" "gene280" "gene848" "gene249" "gene965" "gene877" "gene599" "gene263" "gene607"
## [23] "gene998" "gene17" "gene561" "gene592" "gene123" "gene186" "gene270" "gene872" "gene96" "gene170" "gene645"
## [34] "gene817" "gene239" "gene957" "gene993" "gene222" "gene154" "gene510" "gene983" "gene476" "gene658" "gene253"
## [45] "gene773" "gene973"
##
## attr(,"intersections")$`A:C`
## [1] "gene393" "gene20" "gene71" "gene479" "gene343" "gene826" "gene56" "gene87" "gene8" "gene50" "gene115"
## [12] "gene464" "gene981" "gene760" "gene485" "gene54" "gene255" "gene822" "gene892" "gene359" "gene325" "gene117"
## [23] "gene769" "gene805" "gene497" "gene516" "gene855" "gene488" "gene553" "gene763" "gene862"
##
## attr(,"intersections")$`A:D`
## [1] "gene982" "gene46" "gene901" "gene697" "gene782" "gene635" "gene563" "gene692" "gene269" "gene657" "gene132"
## [12] "gene25" "gene261" "gene211" "gene486" "gene710" "gene991" "gene126" "gene870" "gene121" "gene404" "gene499"
## [23] "gene945" "gene833" "gene172" "gene219" "gene830" "gene406" "gene345" "gene797" "gene444" "gene3" "gene64"
## [34] "gene544" "gene10"
##
## attr(,"intersections")$`B:C`
## [1] "gene703" "gene874" "gene27" "gene627" "gene182" "gene254" "gene41" "gene163" "gene967"
## [10] "gene257" "gene975" "gene899" "gene962" "gene494" "gene845" "gene189" "gene47" "gene326"
## [19] "gene109" "gene940" "gene514" "gene334" "gene588" "gene897" "gene780" "gene264" "gene158"
## [28] "gene424" "gene600" "gene750" "gene587" "gene858" "gene766" "gene511" "gene524" "gene500"
## [37] "gene856" "gene515" "gene861" "gene320" "gene240" "gene338" "gene939" "gene849" "gene810"
## [46] "gene423" "gene775" "gene69" "gene752" "gene622" "gene502" "gene713" "gene542" "gene755"
## [55] "gene431" "gene153" "gene905" "gene598" "gene667" "gene439" "gene192" "gene88" "gene764"
## [64] "gene194" "gene408" "gene282" "gene884" "gene626" "gene162" "gene90" "gene335" "gene995"
## [73] "gene823" "gene273" "gene295" "gene183" "gene468" "gene315" "gene1000" "gene507" "gene348"
## [82] "gene259" "gene803" "gene911" "gene577" "gene1" "gene728" "gene873" "gene223" "gene101"
## [91] "gene63" "gene781" "gene446" "gene864" "gene81" "gene196" "gene195" "gene615" "gene354"
## [100] "gene719" "gene954" "gene549" "gene23" "gene376" "gene613" "gene809" "gene606" "gene26"
##
## attr(,"intersections")$`B:D`
## [1] "gene456" "gene747" "gene839" "gene699" "gene759" "gene364" "gene230" "gene327" "gene886" "gene268" "gene14"
## [12] "gene527" "gene260" "gene112" "gene985" "gene204" "gene681" "gene595" "gene36" "gene128" "gene824" "gene180"
## [23] "gene352" "gene351" "gene971" "gene660" "gene7" "gene224" "gene744" "gene609" "gene793" "gene666" "gene97"
## [34] "gene136" "gene506" "gene285" "gene156" "gene946" "gene725" "gene102" "gene883" "gene33" "gene387" "gene605"
## [45] "gene740" "gene339" "gene329" "gene139" "gene568" "gene432" "gene980" "gene504" "gene505" "gene517" "gene460"
## [56] "gene687" "gene629" "gene218" "gene671" "gene734" "gene375" "gene932" "gene167" "gene639" "gene407" "gene416"
##
## attr(,"intersections")$`C:D`
## [1] "gene399" "gene960" "gene531" "gene708" "gene323" "gene837" "gene704" "gene381" "gene95" "gene458" "gene503"
## [12] "gene493" "gene443" "gene915" "gene281" "gene891" "gene716" "gene636" "gene220" "gene762" "gene179" "gene696"
## [23] "gene844" "gene42" "gene314" "gene184" "gene394" "gene937" "gene896" "gene672" "gene918" "gene103" "gene324"
## [34] "gene721" "gene286" "gene141" "gene928"
##
## attr(,"intersections")$`A:B:C`
## [1] "gene882" "gene157" "gene49" "gene119" "gene384" "gene5" "gene166" "gene807" "gene586" "gene296" "gene969"
## [12] "gene231" "gene108" "gene317" "gene227" "gene934" "gene554" "gene421" "gene316" "gene418" "gene16" "gene330"
## [23] "gene481" "gene714" "gene53" "gene340" "gene730" "gene347" "gene283" "gene181" "gene727" "gene571" "gene836"
## [34] "gene679" "gene445" "gene501" "gene933" "gene410" "gene529" "gene94"
##
## attr(,"intersections")$`A:B:D`
## [1] "gene904" "gene79" "gene207" "gene382" "gene331" "gene753" "gene924" "gene356" "gene142" "gene463" "gene341"
## [12] "gene537" "gene829" "gene144" "gene21" "gene815" "gene4" "gene729" "gene646" "gene931" "gene997" "gene312"
## [23] "gene859" "gene594" "gene465" "gene684" "gene949" "gene539" "gene440" "gene398" "gene425" "gene174" "gene597"
## [34] "gene405" "gene653" "gene612" "gene75" "gene927" "gene378" "gene370" "gene908"
##
## attr(,"intersections")$`A:C:D`
## [1] "gene449" "gene369" "gene78" "gene414" "gene358" "gene749" "gene984" "gene893" "gene37" "gene32" "gene148"
## [12] "gene718" "gene907" "gene138" "gene739" "gene723" "gene578" "gene930" "gene336" "gene680" "gene234" "gene455"
## [23] "gene547" "gene756" "gene388" "gene688" "gene717" "gene754" "gene758" "gene61" "gene552" "gene518" "gene202"
##
## attr(,"intersections")$`B:C:D`
## [1] "gene558" "gene131" "gene238" "gene65" "gene536" "gene955" "gene630" "gene6" "gene641" "gene682" "gene802"
## [12] "gene576" "gene900" "gene827" "gene814" "gene433" "gene624" "gene601" "gene127" "gene228" "gene310" "gene652"
## [23] "gene670" "gene841" "gene894" "gene936" "gene365" "gene711" "gene616" "gene974" "gene566" "gene216" "gene921"
## [34] "gene466" "gene2" "gene302" "gene311" "gene806" "gene520" "gene811" "gene191" "gene277" "gene360" "gene990"
## [45] "gene232" "gene469" "gene400" "gene214" "gene665" "gene72" "gene846" "gene31" "gene92" "gene583" "gene242"
## [56] "gene483"
##
## attr(,"intersections")$`A:B:C:D`
## [1] "gene371" "gene735" "gene787" "gene38" "gene66" "gene415" "gene284" "gene477" "gene768" "gene454" "gene522"
## [12] "gene784" "gene582" "gene34" "gene618" "gene741" "gene850" "gene373" "gene742" "gene912" "gene748" "gene291"
## [23] "gene863" "gene143"
##
## attr(,"class")
## [1] "venn"Conclusion
Cet article décrit de multiples solutions pour créer des diagrammes de Venn dans R.
Version:
 English
English






No Comments